The work investigates the sparse classification methods for high dimensional data. Here I provide a toy example to classify patient with Alzheimer’s Disease (AD) using gene expression data (select 500 gene expression). Two methods were adopted to carry out classification: sparse logistic regression and sparse proximal support vector machine (SVM). I assessed how well the methods can discriminate between patients based on classification accuracy, sensitivity and specificity. I conducted a 5-fold cross validation to avoid overestimation of the performance. All gene expression measures were scaled to zero mean and unit variance. The gglasso package is used. The optimal value of penalty parameter lambda was selected from a series of lambdas which gave the minimum cross validation error on training data set. The simulation results were presented in below tables.
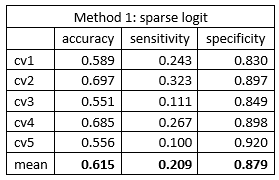
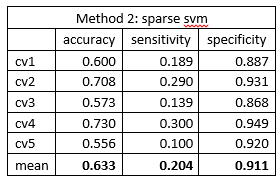
The 500 gene expression gave a relative poor discriminative capability. The mean classification accuracy was around 0.62 via both methods. The sensitivity was surprising low, about 0.2, which indicated a poor capability of identifying AD patients. The high specificity and low sensitivity indicated that the model was inclined to classify patient as cognitive normal. The frequency of cognitive normal patient was 273/447 = 0.611, which was close to the mean accuracies we got from the models. The classification model can be viewed as simple guessing.
I then conducted functional principal component (FPC) analysis on the scaled 500 gene expression data. I selected the first 2 FPC scores which explained 95% of variance in the data. Hotelling’s t-test was conducted between case and control group on FPC scores. The test statistic is 1.49 with p-value 0.226, which indicates that there was on significant different in the gene expression sequence between cognitive normal patients and AD patients. This somehow explains the poor performance of the models in the first analysis.
In conclusion, using all 500 gene expression may not help to identify AD patient. Fitting the model with selected significant genes may improve the discriminative capability. The R code for this toy example is provided.